New genotyping tools reduce costs and improve throughput for wheat breeders, significantly advancing wheat breeding programs in the UK and globally
Submitting Institution
University of BristolUnit of Assessment
Biological SciencesSummary Impact Type
TechnologicalResearch Subject Area(s)
Biological Sciences: Genetics
Agricultural and Veterinary Sciences: Crop and Pasture Production
Summary of the impact
The wheat-breeding industry, including some of the largest plant breeders
and seed-development companies in the world, has benefited from decreased
production costs and increased productivity as a result of research led by
the University of Bristol and carried out between 2009 and 2011. The
Bristol researchers developed the tools necessary to differentiate point
mutations in the complex DNA structure of wheat. This was a critical step
in wheat genotyping and led to the public release of 95% of the wheat
genome in 2010 and the development, by Bristol, of a cheap, easy-to-use
assay for industry. These advances were quickly embraced by industrial
wheat breeders aiming to deliver new varieties of wheat with improved
yields and desirable traits such as disease resistance. Limagrain, the
world's fifth-largest producer of field seeds (including wheat) with €595
million in sales of seeds, realised a ten-fold reduction in costs and
a ten-fold increase in throughputs in their breeding laboratory.
With the wheat-seed business worth over £16 million annually in the UK and
over £1.8 billion globally, the new genotyping tools generated by Bristol
have had, and continue to have, a major impact on the wheat industry and
its ability to respond to the challenges of climate change and population
growth.
Underpinning research
Key researchers and their contributions
Sequencing the wheat genome was a collaborative project between the
University of Bristol, the University of Liverpool and the John Innes
Centre (JIC), with Bristol being the lead organisation. Bristol generated
the cDNA sequence data from the four varieties of wheat studied and used
its expertise in genomics and bioinformatics to decipher the information
and generate the varietal single nucleotide polymorphisms (SNPs). The team
validated the SNPs in a range of germplasm to ensure their suitability for
use by academic and commercial wheat breeders. This research was carried
out between April 2009 and April 2011 by the following key Bristol
researchers:
Professor Keith Edwards (date of appointment 1st January
2001); Chair in Cereal Functional Genomics, School of Biological Sciences
Dr Gary Barker (date of appointment February 2003); Lecturer in
Bioinformatics, School of Biological Sciences
Since April 2011, Bristol has continued to work closely with the breeding
companies to develop SNP markers for use in wheat breeding.
Context
SNPs are regions of DNA, between individual chromosomes, that vary by a
single nucleotide. They are the most common type of genetic variation in
plant and animal genomes. For this reason, SNPs are a logical choice for
marker-assisted selection — a process used in breeding programmes
selecting for traits such as disease resistance and productivity. However,
the wheat genome is extremely complex: five times larger than the human
genome, the hexaploid wheat genome has three sets of chromosomes derived
from related progenitors. This has presented significant challenges in SNP
discovery, as work by the Bristol group has shown that most SNPs
identified represent sequence differences between the three sets of
chromosomes within an individual variety rather than between chromosomes
from different varieties [1]. As a result, the Bristol team had to develop
and adapt a range of novel procedures in the area of next-generation
sequencing, bioinformatics and genotyping in order to overcome these
challenges and advance DNA marker-assisted breeding approaches.
Nature of research findings
SNP genotyping first requires the identification of valid SNPs. The
Bristol team used a bioinformatics approach to electronically mine SNPs
from several databases containing expressed sequence tags (ESTs) for
cereals, including wheat. Their findings showed that high-throughput
sequencing could generate a dataset of sufficient size to exclude SNPs
within a variety from those found between varieties yet still produce
sufficient SNPs to provide diverse genome coverage [2]. Simultaneously,
the Bristol team was exploring high-throughput sequencing methods and
developing an assay, based on novel technologies (for instance, padlock
probes and Kompetitive Allele Specific PCR (KASP)), capable of
simultaneously genotyping numerous wheat lines using several hundred
gene-based SNP markers [3]. This work paved the way for the sequencing and
genotyping of the wheat genome.
In 2010, using the wheat reference variety Chinese Spring, line 42,
Bristol employed novel bioinformatics techniques to generate SNPs among
the sequence information covering 95% of the wheat genome [4]. The
sequence data were made available immediately in the public domain through
the EMBL/GenBank and CerealsDB [5] websites, which allowed rapid uptake of
this information by both the academic and commercial wheat-breeding
communities.
To make the genomic sequence information accessible to industry and
assist practical breeding of wheat, the Bristol team combined their
bioinformatics expertise with the innovative KASP chemistry developed by
the genetic-screening company, KBioscience (now LGC), to generate and
validate a new type of wheat molecular marker (KASP markers) [6]. The
markers are cheap and easy to use, which makes them ideal for commercial
wheat-breeding programmes aimed at the fast development of new varieties
selected for increased yield and resistance to abiotic and biotic stress.
References to the research
Outputs
[1] Barker, G.L.A. & Edwards, K.J. (2009) `A genome-wide analysis of
single nucleotide polymorphism diversity in the world's major cereal
crops', Plant Biotechnology Journal, 7: 312-317. DOI:
10.1111/j.1467-7652.2009.00412.x.
[2] Edwards, K.J., et al. (2009) `Multiplex SNP-based genotyping
in allohexaploid wheat using padlock probes', Plant Biotechnology
Journal, 7: 375-390. DOI: 10.1111/j.1467-7652.2009.00413.x.
[3] Allen, A.M., et al. (2011) `Transcript-specific,
single-nucleotide polymorphism discovery and linkage analysis in hexaploid
bread wheat (Triticum aestivum L.)', Plant Biotechnology
Journal, 9: 1086-1099. DOI: 10.1111/j.1467-7652.2011.00628.x.
[5] Brenchley, R., et al. (2012) `Analysis of the allohexaploid
bread wheat genome using whole genome shotgun sequencing', Nature,
491: 705-710. DOI: 10.1038/nature11650.
[6] Allen, A.M. et al. (2013) `Discovery and development of
exome-based, co-dominant single nucleotide polymorphism markers in
hexaploid wheat (Triticum aestivum L.), Plant Biotechnology
Journal, 11 (3): 279-95. DOI: 10.1111/pbi.12009.
Grants
[7] Edwards (2009-2011) Mining the allohexaploid wheat genome for
useful sequence polymorphisms, BBSRC, £1.2 million (£250,000 to
Bristol; Edwards lead PI).
[8] Edwards (2011-2013) Development and validation of a flexible
genotyping platform for wheat, BBSRC Crop Club, £200,000 (Edwards
lead PI).
Award
In 2011, Professor Edwards was awarded the Royal Agricultural Society of
England's (RASE) Research Medal for this research. The RASE Research Medal
acknowledges "research work of outstanding merit carried out in the UK,
which is proven or likely to be of benefit to agriculture" [a].
Details of the impact
In August 2010, a consortium funded by BBSRC and led by the research team
at the University of Bristol released genomic sequences covering 95% of
the wheat genome through the EMBL/GenBank and CerealsDB [5] websites. This
empowered wheat breeders to carry out wheat genotyping: they could begin
to sequence genes of interest, such as those related to growth or disease
resistance, by analysing target DNA sequences against the database
supplied by the University of Bristol [b, c]. These data have been
extensively accessed, with the CerealsDB website (http://www.cerealsdb.uk.net/)
receiving the following numbers of visits between 2009 and 2012:
|
2009 |
2010 |
2011 |
2012 |
2013
(to 31/05) |
| Total |
45,511 |
67,269 |
199,139 |
196,303 |
112,946 |
Evidence that the sequences available on CerealsDB have international
impact can be found by examining the locations of those accessing the
site's content:
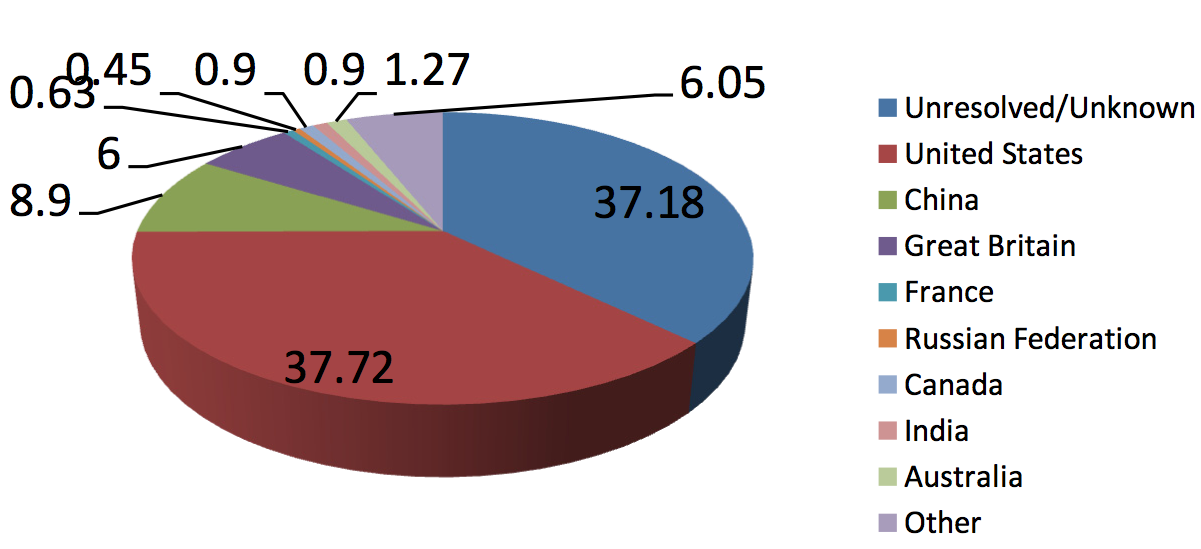 Figure 1: Summary of the locations of the users of CerealsDB (during 2012)
Figure 1: Summary of the locations of the users of CerealsDB (during 2012)
Research leads to new commercial genotyping service for KBioscience
For sequence information to be useful to wheat breeders, a cost-effective
method of analysis and a large number of SNP markers were needed. To
achieve this, the Bristol group developed novel procedures to deal with
the complex DNA of wheat [2,3,4] and then collaborated with the genetic
screening company, KBioscience, which had developed the KASPar assay
[6].This collaboration enabled KBioscience to offer a new commercial
genotyping service to the global wheat community. Though the number of
screenings carried out using the wheat SNPs is commercially sensitive, the
Principal Scientist at KBioscience said: "The markers are used extensively
by the global wheat community and they constitute a significant part of
the KBioscience portfolio" [d]. The commercial success of this new service
"continues to provide benefits to the UK economy in terms of the
genotyping services that KBioscience offer and via the employment of high
quality technicians and scientists based on our ability to offer this
service" [d].
Genotyping tools lead to decreased production costs and increased
productivity for the wheat-breeding industry
The UK wheat-seed business is worth £16 million per annum, with each ton
of seed having a profit value of £56-80 depending on the variety. One ton
of wheat seed can plant ~7 hectares. Globally, the wheat-seed business is
worth over £1.8 billion a year. The farm-gate value of wheat is £180 per
ton, or £2.9 billion for the whole of the UK. The new genotyping tools
generated by Bristol have had a major impact on how the wheat community
carries out its genotyping, resulting in decreased costs and increased
productivity for a multi-billion pound industry [b,c,e,f].
Limagrain is the largest plant breeder and seed-development company in
Europe and the fifth-largest field-seed producer in the world. Field-seed
sales are one of their three main activities and represent 38% of their
global sales (over €1.5 billion in 2011) [g, pg. 9]. Prior to the research
developments at Bristol, Limagrain was severely hampered in its objective
of switching over to using SNPs, despite knowing they were the more
efficient molecular marker for use in plant breeding. The research at
Bristol enabled Limagrain to change from Simple Sequence Repeat (SSR)
markers to SNP markers, allowing a 10-fold reduction in the cost per
data point, from €0.50 to €0.05, due to increased automation,
reduction in reagent use and improved accuracy and ease of interpretation
[c]. The switch to SNP genotyping also significantly improved efficiency.
The Head of Research — Cereals at Limagrain stated that "the throughput
of the Limagrain Laboratory was increased ten-fold" as work that
took three months using SSRs took just three days using the SNP markers
[c].
RAGT Seeds is another one of Europe's leading seeds businesses, with
wheat being one of their key crops. The advances made by Bristol have
become "embedded in [RAGT's] systems" and are considered "indispensable
tools" [e]. They have led to a five-fold increase in efficiency
in the RAGT laboratory, which in turn has resulted in a doubling of
the rate of genetic gain for traits such as yield, disease resistance and
quality [e].
KWS, one of the leading cereal breeders in the European Union, has also
benefited from this research, realising a ten-fold increase in
marker-screening capability and recently expanding their development
team by two people in their cereals division [f].
Reaching beyond the UK
All of the companies that have benefited from these advances in wheat
genotyping have operations that extend beyond the UK. For example, KWS
operates in 70 countries worldwide, while the Limagrain group is the fifth-largest
plant breeder and seeds company in the world, employing over
6,000 people and turning over more than £1.1 billion annually.
Professor Edwards and Dr Barker and their team have consciously
coordinated their research efforts with other international and UK
programmes. Specifically, the SNPs identified within the project have made
a significant contribution to international efforts to develop chip
platforms, which offer commercial breeders an alternative assay platform
for establishing large-scale patterns (F).
Significance of the impact
The wheat-breeding industry has benefited from this research since 2010,
helping it to realise considerable improvements in productivity and
reductions in costs. These commercial benefits have already stimulated
further international investment in projects that aim to improve our
knowledge and consequently our ability to exploit the genetic diversity of
wheat to improve yields in the face of a growing global population and
environmental change. For example, BREEDWHEAT is an international
consortium of 26 partners, including industry partners such as Limagrain,
with a €39 million investment programme over nine years [g, pg 23]. In
2011, BBSRC awarded a £7 million grant to a consortium of researchers,
including Edwards, that aims to identify new and useful genetic traits
that will improve modern wheat-breeding efforts. This collaborative work
is being undertaken around the globe to try and address concerns over food
security. Though considerable work remains in bringing the genomic
sequencing of wheat up to the level of information that is currently
available for crops such as rice, maize and soybeans, the contributions
made by the researchers at Bristol represent significant advances, the
impacts of which will continue to grow as new varieties of wheat are
developed.
Universities and Science Minister David Willetts said that the work
conducted at the University of Bristol was "... an outstanding world class
contribution by the UK to the global effort to completely map the wheat
genome. By using gene sequencing technology developed in the UK we now
have the capability to improve the crops of the future by simply
accelerating the natural breeding process to select varieties that can
thrive in challenging conditions." [h]
Sources to corroborate the impact
[a] Royal Agricultural Society of England (2012). What the Royal
Agricultural Society of England Does — Awards. <http://www.rase.org/what-we-do/awards/>
[accessed online 4 June 2013].
[b] Head of Laboratory, Syngenta.
[c] Head of Research — Cereals, Limagrain.
[d] Principal Scientist, KBioscience.
[e] Cereal Genotyping Lead, RAGT Seeds.
[f] Barley Pre-Breeder, Cereals Division, KWS UK Ltd.
[g] Limagrain (2011) From earth to life: Limagrain Annual Report 2011.
<http://www.limagrain.com/docs/fckeditor/file/publications/RA/Lmg_RA2011_GB.pdf>
[h] BBSRC. (2010) UK researchers release draft sequence coverage of
wheat genome. [Press release 27 August 2010]. <http://www.bbsrc.ac.uk/news/food-security/2010/100827-pr-uk-researchers-release-draft-wheat-genome.aspx>