UOA10-03: Pharmaceutical and biotechnology companies gain economic benefits from novel statistical methods for imputing genotypes
Submitting Institution
University of OxfordUnit of Assessment
Mathematical SciencesSummary Impact Type
TechnologicalResearch Subject Area(s)
Mathematical Sciences: Statistics
Biological Sciences: Genetics
Summary of the impact
In genetic studies of human disease it is now routine for studies to
collect genetic data on
thousands of individuals with and without a particular disease. However,
the genetic data collected
is incomplete, with many millions of sites of the genome unmeasured. The
novel methods and
software (IMPUTE) developed by researchers at the University of Oxford
predict unobserved
genetic data using reference datasets.
IMPUTE has been adopted by the company Affymetrix in the design of custom
genotyping chips.
Affymetrix recently won the tenders by the UK Biobank and UKBiLEVE studies
to genotype
>500,000 participants, with a total study cost of ~£25M. The company
states that IMPUTE gave
their project bid a significant competitive advantage. Affymetrix also
purchased the IMPUTE source
code for £250,000. In addition, Roche Pharmaceuticals have used the
software in their research on
the genetic basis of drug response. The use of imputation has saved Roche
~$1,000,000.
Underpinning research
Genome-wide association studies (GWAS) aim to identify genes that
increase risk of developing a
disease under study. A typical study will measure up to a million variable
positions across the
genome, called single nucleotide polymorphisms (SNPs), in thousands of
subjects, and look for
significant differences between individuals with and without the disease.
The identification of these
disease genes can help understanding of the disease mechanisms. Since only
a fraction of sites
that are known to vary between humans are measured, there is a substantial
amount of genetic
data that is unobserved. However, reference databases such as the 1000
Genomes Project (TGP)
contain many more SNPs. The July 2012 TGP release contains 38 million
SNPs. The methodology
developed at the University of Oxford combines the data from a GWAS with
the TGP database and
predicts the unobserved genotypes.
The first approach at predicting, or imputing, unobserved genotypes was
developed by Dr Marchini
and Professor Donnelly, both faculty members at the University of Oxford,
as part of their
involvement in the Wellcome Trust Case Control Consortium (WTCCC) [1],
during the period of
2006-2007. They realized that genetic studies of human disease could be
substantially improved if
unobserved genotypes could be predicted using the existing reference
databases, and that
recently developed Hidden Markov models developed in the area of
population genetics could be
adapted to carry out this task. Their approach, IMPUTE v1 [2], was
developed by Marchini and
applied successfully to all 7 disease studies carried out by the WTCCC.
This paper has over 1,000
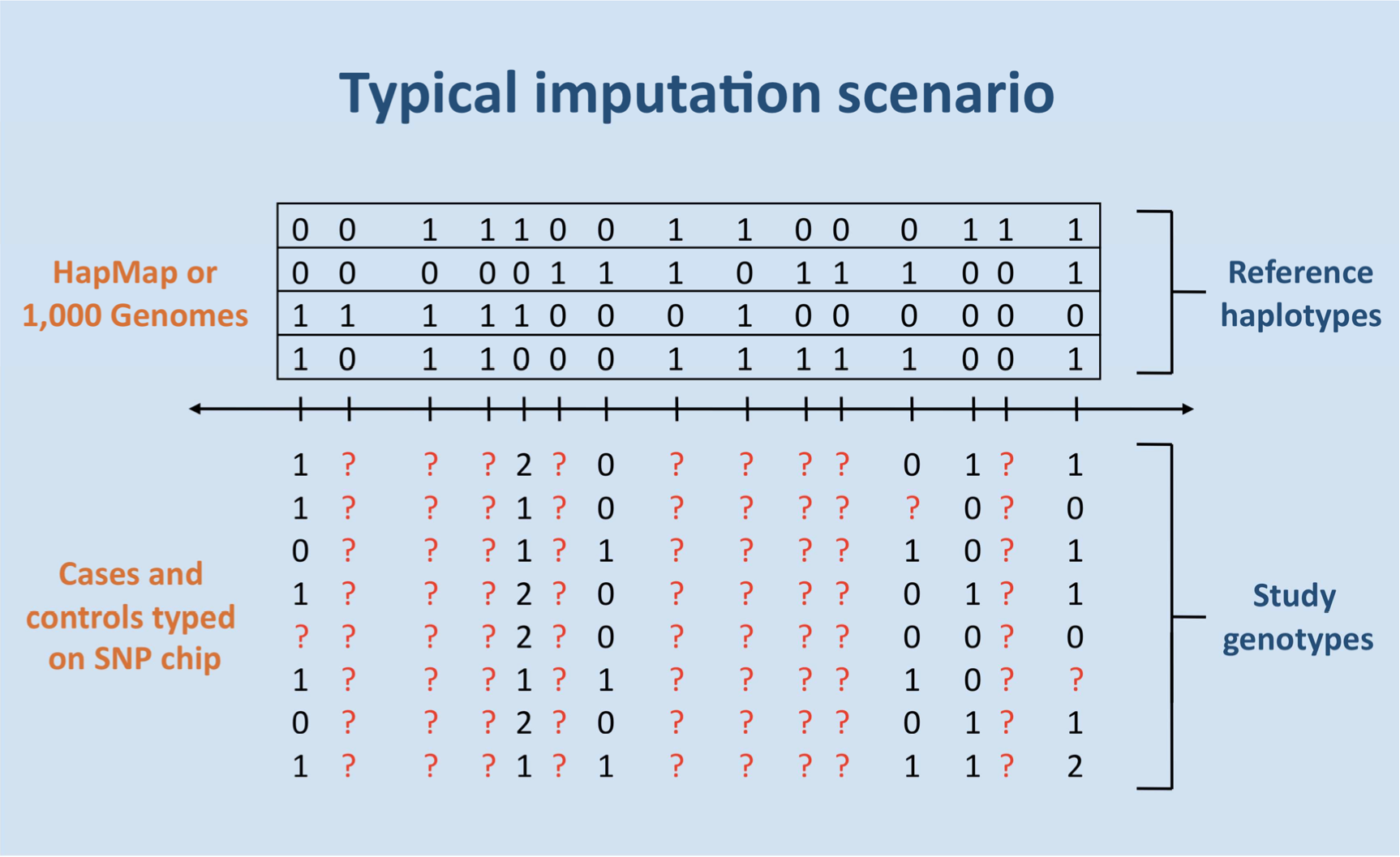
citations since 2007. The figure below illustrates the typical imputation
scenario, where a reference
panel of haplotypes is combined with a GWAS. The figure highlights that a
large fraction of
genotypes are unobserved (indicated by question marks). IMPUTE can predict
this missing data
using shared patterns of haplotypes between the two datasets. For common
genetic variants of
interest, the accuracy of imputation is over 95%.
There have been over 1,350 published GWAS since 2005 (www.genome.gov/gwastudies).
Imputation
has been used in the vast majority of these, evidenced by the large number
of citations
of our papers on imputation. One key benefit of the method is that once
unobserved genotypes
have been predicted in several different studies, they can then be
combined, via meta-analysis, to
produce much more powerful studies. This approach has changed the field of
human genetics and
groups now routinely share data via this approach. One of the earliest
examples of this was in the
study of Type 2 Diabetes and lead to the discovery of 6 new disease genes
[3].
Subsequently, Marchini and Donnelly realized that as reference panels
increase in size, through
ongoing projects such as the TGP, the method IMPUTE v1 would not scale
well. Marchini led the
development of IMPUTE v2 which extends the approach by adaptively
selecting a subset of the
reference database to use for predicting each individual. Another insight
was that this approach
naturally allows the use of reference panels from multiple populations.
For example, when
predicting genotypes in an individual with European ancestry the method
would select the subset
of the reference database that matches the individual's ancestry [4,5].
A further paper published in Nature Genetics [6] develops a new two-step
imputation process, first
by estimating haplotypes in the GWAS sample, then using haploid
imputation. The second step is
very fast and reduces the computational cost needed by a factor of at
least 20.
From 2002-2005, Marchini held a Wellcome Trust Postdoctoral Fellowship at
the University of
Oxford and since 2005 has been a University Lecturer in Statistical
Genomics. Donnelly has been
a Professor of Statistical Science since 1996. From 2007 he has also been
Director of the
Wellcome Trust Centre for Human Genetics, University of Oxford.
References to the research
*[1] The Wellcome Trust Case Control Consortium (2007) Genomewide
association study of
14,000 cases of seven common diseases and 3,000 shared controls. Nature
447 661-78.
doi:10.1038/nature05911.
*[2] J. Marchini, B. Howie, S. Myers, G. McVean and P. Donnelly (2007) A
new multipoint method
for genome-wide association studies via imputation of genotypes. Nature
Genetics 39 906-
913. doi:10.1038/ng2088.
[3] E. Zeggini, L. Scott, R. Saxena, B. Voight, J. Marchini et al. (2008)
Meta-analysis of genome-
wide association data and large-scale replication identifies additional
susceptibility loci for
type 2 diabetes. Nature genetics 2008;40;5;638-45.
doi:10.1038/ng.120.
*[4] B. Howie, P. Donnelly, J. Marchini (2009) A Flexible and Accurate
Genotype Imputation
Method for the Next Generation of Genome-Wide Association Studies. PLoS
Genetics 5(6):
e1000529. doi:10.1371/journal.pgen.1000529.
[5] B. Howie, J. Marchini, M. Stephens (2011) Genotype Imputation with
Thousands of
Genomes. G3 : Genes, Genomes, Genetics. doi: 10.1534/g3.111.001198.
[6] B. Howie, C. Fuchsberger, M. Stephens, J. Marchini, and G. R.
Abecasis (2012) Fast and
accurate genotype imputation in genome-wide association studies through
pre-phasing.
Nature Genetics 44, 955-959. doi: 10.1038/ng.2354.
The three asterisked outputs best indicate the quality of the
underpinning research. All six papers
are in high quality internationally refereed journals.
Details of the impact
There are two main areas where IMPUTE software has made an economic
impact on companies
working in the area of genetics and pharmaceuticals:
- IMPUTE has had a significant impact on the company Affymetrix. It has
led to the
introduction of new products and has significantly changed a design
process. The company
has benefited by recently winning a genotyping contract worth ~£25M.
- IMPUTE has led to the improvement of a drug response study carried out
by Roche. This
saved the company an estimated ~$1,000,000.
Affymetrix licensed the source code for both IMPUTE v1 (2009) and v2
(2010) from Oxford
University for £250,000 [A]. Affymetrix use Impute v2 as a central part of
the process of designing
both generic and custom-made SNP chips (a chip is a collection of
microscopic DNA spots
attached to a solid surface). In addition, licences for use of the
software, without the source code,
worth ~£70,000 in total have been sold to Genentech (2008),
GlaxoSmithKline (2008),
Biocomputing Platforms Ltd. (2009) and PGxHealth (2010) [A]. IMPUTE has
also been used in a
study of drug response by Roche via a 2011 consultancy agreement with
Marchini.
Optimizing product design at Affymetrix using IMPUTE
Genotype imputation is now a central method in human genetics utilized by
researchers carrying
out GWAS. The method is usually applied to data collected from genome-wide
SNP arrays.
Affymetrix is a $300M US company that makes such arrays together with the
equipment and
reagents to run the experiments. Such equipment is essential in any lab
carrying out its own
GWAS.
The company has used IMPUTE in the design process for a new series of
population-specific
arrays called the "AxiomTM Genome-Wide EUR, EAS, LAT and AFR Arrays",
targeted at the
European, East Asian, Latino and African populations. These arrays are
sold commercially to
research groups carrying out GWAS [B,C]. Affymetrix recently won the bids
to genotype >500,000
participants for the UKBiLEVE (http://www.mrc.ac.uk/Newspublications/News/MRC008925)
and
UK Biobank (http://www.ukbiobank.ac.uk/)
studies, with a total study cost of ~£25M. The UK
Biobank project is the largest single genotyping study on record in the
world as well as the largest
single project in the Affymetrix Genotypic revenue base [B]. The company
states that "a significant
competitive advantage of the Affymetrix proposal was a custom GWAS grid
that draws significant
power from using IMPUTE2 in its design" [D].
The Vice President for Informatics at Affymetrix says in [B] "The
impute software that we licenced
from Oxford University has been used extensively at Affymetrix and is an
essential tool used to
compute and describe the coverage of our genotypic arrays. [...]
In particular, the SNPs on these
arrays were selected in such a way as to maximize imputation coverage.
This has made a
significant impact in the way we design arrays and could not have
happened without using
IMPUTE2, which has been shown to be the most accurate method
of imputation in the literature.
[...] Affymetrix had total revenues of about $300 million in 2012 and
is using IMPUTE2 in the
design and dissemination of all its genotyping products Affymetrix has a
significant and rapidly
growing share of the worldwide market for genotyping arrays, the size of
which is on the order of
$600m million annually."
Roche saved ~$1,000,000 by using IMPUTE in a study of drug response
Pharmacogenetics is a particular type of GWAS applied to subjects that do
or do not have an
adverse reaction to a particular medication. Many medications exhibit a
variable response rate that
is thought to be partly genetic. Therefore, there is a great interest in
discovering biomarkers that
aid physician decision making, through the identification of patients who
will or will not respond,
and therefore derive greater benefit from a particular therapy.
The pharmaceutical company Roche has investigated the genetics of
response to the drug
tocilizumab for the treatment of rheumatoid arthritis (RA). Tocilizumab is
prescribed to RA patients
who had inadequate response to disease modifying anti-rheumatic drugs.
Genotype imputation
using IMPUTE v2 was used in this study to combine studies together for
greater power. Since the
subjects in these studies had a variety of different ancestries the use of
IMPUTE v2 together with
the HapMap3 reference panel provided an ideal and practical solution to
the prediction of the
unobserved genotypes in each study. The study was able to implicate the
involvement of 8 loci in
the patient response to tocilizumab treatment. Patients carrying the
specific genetic markers had a
higher remission compared to those who did not [F].
The Roche study used three different Illumina genotyping chips (550K,
Human1M-Duo and
HumanOmni1-Quad) on different sets of individuals. A Senior Statistical
Scientist at Roche [E]
states "The IMPUTE program was used and generated high quality data for
the union set of SNPs
on the three chips. This allowed us to analyse the data from all 1600
patients together.....Without
the genetic data imputation carried out with IMPUTE, the best way to
reproduce the study would be
to genotype all study samples using an IlluminaOmni1-Quad chip. This
would have involved re-
genotyping 1,157 samples at a cost of $750 each plus an additional
operational cost of 20%.
Therefore the total cost saving is ~$1,000,000. In addition to the cost
saving, the imputation work
also allowed us to save time and complete the analysis in time to meet
decision timelines set by
the development program".
Sources to corroborate the impact
[A] Letter from Technology Transfer Team Leader, ISIS Innovation, Oxford,
held by the
University of Oxford, which corroborates licensing deals and software
sales for IMPUTE.
[B] Letter from Vice President Informatics, Affymetrix, held by the
University of Oxford, which
corroborates how Affymetrix have made use of IMPUTE.
[C] Affymetrix press release giving details of their Axiom arrays and how
IMPUTE was used to
design the arrays, copy held by the University of Oxford
[D] Affymetrix press release giving details of contract with UK Biobank,
copy held by the
University of Oxford
[E] Letter from Senior Statistical Scientist, Roche, held by the
University of Oxford, describing
the use of IMPUTE in their pharmacogenetic study of Tocilizumab for the
treatment of
rheumatoid arthritis.
[F] Paper describing Roche's pharmacogenetic study of Tocilizumab,
confirming the use of
IMPUTE in their study.
Wang J et al. (2011) Genome-wide association analysis implicates the
involvement of 8 loci
with response to tocilizumab for the treatment of rheumatoid arthritis The
Pharmacogenomics
Journal 44, 955-960, doi: 10.1038/tpj.2012.8