Impact on DNA (gene) sequencing based on chemically modified DNA
Submitting Institution
University of LiverpoolUnit of Assessment
ChemistrySummary Impact Type
TechnologicalResearch Subject Area(s)
Biological Sciences: Biochemistry and Cell Biology
Summary of the impact
This case study describes both economic and healthcare benefits that have
resulted from a new DNA (gene) sequencing technique known as SOLiD
sequencing. Through the 1990s until the present, Cosstick (University of
Liverpool since 1984) has both developed the synthesis and studied the
properties of chemically modified DNA in which a single oxygen atom is
replaced by sulfur; we have termed this a 3'-phosphorothiolate (3'-sp)
modification. Chemically prepared DNA containing the 3'-sp modification is
a key enabling component of the Applied Biosystems SOLiD DNA sequencing
instrument which is able to produce extremely rapid, cost-effective and
exceptionally accurate DNA sequence information. The impact of this very
powerful sequencing technology extends beyond economic benefits as it has
many healthcare applications which have impacted medical practice.
Underpinning research
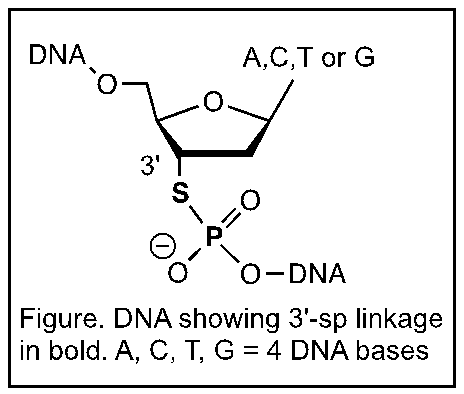
The chemical modification to DNA that is required for the SOLiD
sequencing technology involves replacing a single oxygen atom in the
phosphodiester linkage of the DNA with sulfur to produce the
3'-phosphorothiolate (3'-sp) linkage (Figure). This modification was
originally designed by Cosstick (then lecturer in Chemistry, University of
Liverpool) as a DNA analogue that was expected to be resistant to cleavage
by enzymes that process and manipulate DNA, but susceptible to cleavage by
mild chemical reagents. The motivation for this research was to provide
new approaches to manipulate DNA that might find applications in
biotechnology. The chemical synthesis of oligonucleotides (short pieces of
DNA) containing this modification was first reported by the Cosstick
Group. An important step towards realising the potential application of
the 3'-sp modification was the subsequent report by Cosstick and
co-workers that a DNA molecule with 7250 natural phosphate linkages could
be specifically cleaved at the site of a single 3'-sp linkage by aqueous
solutions of silver ions.1 It was apparent at this time that in
order to fully explore the potential applications of the 3'-sp
modification greatly improved methods were required for its preparation
and thus through the 1990s and 2000s the Cosstick group reported a number
of methods for the chemical synthesis of DNA sequences containing the
3'-sp modification. These studies culminated in the key report (2004) of a
preparative method based on the use of 3'-S-phosphorothioamidites
which was compatible with standard automated procedures for the chemical
synthesis of DNA. This method therefore made oligonucleotides containing
the 3'-sp linkage much more readily accessible by routine DNA synthesis
procedures.2
The resulting improved access to these 3'-sp DNA analogues made it
possible to investigate their interactions with complementary sequences of
DNA or RNA in much greater detail. It was shown that a DNA strand
containing this modification could form duplex structures with either
complementary DNA or RNA strands and indeed the 3'-sp modification
actually stabilised some of these structures.3,4 Significantly,
it was established that the effect of the 3'-sp modification had a
predictable effect on the thermal melting temperature of complementary
oligonucleotides. This was important in establishing that DNA containing
the 3'-sp modification would be compatible with the hybridisation steps in
the SOLiD sequencing technique.
In summary, the work conducted by the Cosstick group at Liverpool has
covered the synthesis of the 3'-sp DNA analogues, the effect that this
modification has on duplex and higher order DNA/RNA structures and their
mild chemical and enzymatic cleavage. This work established information on
all the important chemical properties of this modification and thus
provided the necessary background from which the chemical/biochemical
aspects of the sequencing method could be developed. Much of this work was
performed though national and international collaborations. The work has
been supported by both the EPSRC5 and the BBSRC.5
References to the research
* = Three references to indicate the quality of the research.
1. Vyle, J.S., Kemp, D., Cosstick, R., and Connolly, B.A. Sequence and
strand specific cleavage in oligonucleotides and DNA containing
3'-thiothymidine. Biochemistry 31, 3012 - 3018 (1992).
DOI: 10.1021/bi00126a024
2*. Sabbagh, G., Fettes, K.J., Gossain, R., O'Neil, I.A. and Cosstick, R.
Synthesis of phosphorothioamidites derived from 3'-thio-3'-deoxy-thymidine
and 3'-thio-2',3'-dideoxycytidine and the automated synthesis of
oligodeoxynucleotides containing a 3'-S-phosphorothiolate linkage.
Nucl. Acids Res. 32, 495-501 (2004). DOI:
10.1093/nar/gkh189
3*. Beevers, A.P.G., Fettes, K.J., Roberts, S.M., O'Neil, I.A., Arnold,
J.R.P., Cosstick, R. and Fisher, J. Probing the effect of a 3'-S-phosphorothiolate
link on the conformation of a DNA:RNA hybrid; Implications for antisense
drug design, J. Chem. Soc., Chem.Commun. 1458-1459 (2002). DOI:
10.1039/B203582K
4*. Bentley, J., Brazier, J.A., Fisher, J. and Cosstick, R. Duplex
stability of DNA·DNA and DNA·RNA duplexes containing 3'-S-phosphorothiolate
linkages. Org. Biomol. Chem. 5, 3698-3702 (2007). DOI:
10.1039/b713292a
5. Cosstick PI on all grants. EPSRC: EP/F011938/1, Increasing the potency
of RNA interference using RNA mimetics, 2007-10, £299,576; BBSRC: B05097,
Synthesis and application of nucleic acid analogues containing
3'-S-phosphorothiolate linkages, 1995-98, £136,725; BBSRC: B18146,
Oligonucleotides containing phosphorothiolate linkages, 2003-06, £192,736.
Details of the impact
The body of research published by the Cosstick group from the 1990's
through to the present, has described a wealth of information relating to
the chemistry and biochemistry of the 3'-sp linkages. Oligonucleotides
containing this 3'-sp linkage are an essential component in a DNA
sequencing method known as SOLiD™ (Sequencing by Oligonucleotide Ligation
and Detection). SOLiD sequencing was launched by Applied Biosystems Inc
(ABI, now incorporated into Life Technologies as of 2008) in 2007 and the
SOLiD sequencing instruments became commercially available in 2008.6,7
The SOLiD method is based on sequencing by ligation and uses universal
sequencing primers (essentially short oligonucleotides containing the 3-sp
linkage) to interrogate the DNA template to be sequenced. The sequence is
read through rounds of hybridisation, ligation, detection and cleavage.
The cleavage step removes the fluorescent label (required for the
detection step) from the 5'-end of the oligonucleotide and resets the
system for the next round of hybridisation and ligation, so that the next
nucleotide can be sequenced. The specific cleavage step of the 3'-sp
linkage, which is conducted under mild conditions compatible with the
requirements of the SOLiD method using aqueous silver ions as demonstrated
by Cosstick,1 is crucial: at the time when the method was
developed, no other of DNA modification was known to work in SOLiD
sequencing. Details of the sequencing method and the role of the 3'-sp
linkage are clearly evident from the patents of McKernan8 and
from a review article on sulfur analogues of nucleic acids by Zon.9
Cosstick's chemistry was thus decisive in enabling implementation of SOLiD
sequencing. It was subsequently shown that SOLiD sequencing can also be
performed in what is known as "the reverse direction" using the isomeric
5'-sp linkages,8,9 but in this case an additional step is
necessary to remove the 3'-phosphate prior to ligation.
In 2006, ABI were working on the development of their SOLiD sequencing
method and needed to produce oligonucleotides containing the 3'-sp
linkages on a large-scale, as these were essential reagents for the SOLiD
instruments. Their synthetic approach to the large-scale synthesis of
these oligonucleotides was principally based on scaling-up the synthetic
procedures published by Cosstick in 2004.2 On invitation from
Dr Gerry Zon (then director of sequencing chemistry), Cosstick visited the
ABI chemistry labs (Foster City, CA, USA) in October 2006 for detailed
discussions relating to some of the difficulties the chemistry sequencing
group had encountered with the synthesis of 3'-sp modified
oligonucleotides. At that time there was considerable urgency to establish
an efficient method for the production of the 3'-sp modified
oligonucleotides in order to meet the demand for these reagents when the
instruments became commercially available.
In terms of their performance, the latest SOLiD instruments (5500 series
genetic analysers) deliver greater than 90 giga base-pairs of sequence
information in one day and a 7 day run can complete the sequence of the
human genome. The two-base encoding, which is unique to the SOLiD method,
provides exceptional sequencing accuracy at >99.94%, (for
further information see: http://tools.lifetechnologies.com/content/sfs/brochures/cms_057511.pdf),10
as each nucleotide in the sequence is essentially read twice. The SOLiD
technology is one of several so-called second/next-generation sequencing
systems (together with instruments from Illumina, Roche and to a lesser
extent Helicos) which were available as of 2008. Each of these sequencing
systems seems to have its own advantages and limitations, although in 2011
the SOLiD system was reported to have the lowest reagent cost needed
to reassemble a human genome11 and came top in the J.P.
Morgan Next Generation Sequencing Survey12 (published in 2010)
for accuracy. Accuracy was also shown to be the most important attribute
when choosing a DNA sequencing system.12
The estimated sales value of SOLiD sequencing systems to Life
Technolgies is also presented in the J.P. Morgan survey12
and rose from zero in 2007 to $68 million in 2008, $100million in 2009
and predicted sales of $136.9 million in 2010, $178 million in 2011 and
$222.5 million in 2012 (figures based on J.P. Morgan estimates and
company reports12). In 2010, only two years after becoming
commercially available, the SOLiD instrument was estimated to have about
20% market share for second/next-generation sequencing and was predicted
to rise to 22.6% (second largest market share after Illumina) by 2012.12
New jobs created by commercialization of SOLiD sequencing include
USA-based manufacturing of consumable reagents, world-wide hiring of
technical specialists to support customers, and (especially) world-wide
sample preparation, sequencing and bioinformatics analysis at
service-provider facilities.
The benefit of the SOLiD sequencing system goes beyond that of generating
economic impact, as the technology is now enabling patients to benefit
from personalised medicine derived from DNA sequence information.
For example, whole genome sequencing (conducted with the SOLiD 4
instrument) of twins with dopamine-responsive dystonia, a clinically
complex neurological movement disorder, that is normally treated with
L-dopa, identified unexpected mutations in the gene encoding sepiapterin
reductase. This enzyme is responsible for the synthesis of co-factors
required for the synthesis of dopamine and seratonin. Supplement of the
L-dopa therapy with a serotonin precursor resulted in considerable
clinical improvements in both twins.13
The exceptional accuracy of the 2-base encoding means that SOLiD
sequencing is ideally suited to cancer research/diagnosis, because tumors
contain mixed sub-populations with different mutations. Deep-sequencing on
the SOLiD platform has revealed differential expression of microRNAs in
favourable versus unfavourable neuroblastoma (the most common extracranial
solid tumor of childhood) and provides a reliable method to assess the
aggressiveness and hence prognosis of the tumor.14 Whilst just
two specific examples are presented here, they indicate the tremendous
scope of the SOLiD sequencing technique to aid diagnosis and treatment of
a wide range of diseases.
There is a list of over 100 publications resulting from SOLiD sequencing
at the Life Technologies website which cover applications on: sequencing
accuracy, bioinformatics, de novo sequencing, targeted resequencing and
transcriptome analysis (further details available at: http://www.invitrogen.com/site/us/en/home/Products-and-Services/Applications/Sequencing/Next-Generation-Sequencing/Publications-Literature.html
)10
The wealth of research that was conducted by the Cosstick group on DNA
containing the 3'-S-phosphorothiolate linkage has been absolutely
crucial to the development of SOLiD sequencing. The contribution that the
work in Liverpool made to the SOLiD programme has been corroborated by Dr
Gerry Zon15 (letter available) who was directing the ABI
chemistry sequencing group in Foster City (USA) at the time SOLiD
sequencing was being developed. To quote from Dr Zon's letter, "the
scale-up of the synthesis of the phosphorothiolate-containing DNA
reagents had to be done in a very short period of time, because of
competing sequencing techniques which were due to come on the market. It
was an enormous benefit therefore to be able to use and adapt your
published procedures, particularly the automated synthesis of the
phosphorothiolate oligomers you published in 2004" (reference 2). To
quote further, "from the work you had published (reference 3 is
quoted in the letter, it was apparent that) the phosphorothiolate
reagents would be fully compatible with all aspects of the sequencing
technique (and) gave ABI confidence to commit to the technique".
Sources to corroborate the impact
-
http://en.wikipedia.org/wiki/ABI_Solid_Sequencing10
- Metzker, M. Application of next-generation sequencing. Nature
Reviews Genetics 11, 31-46 (2010). DOI: 10.1038/nrg2626
- McKernan, K., Blanchard, A., Kotler, L. & Costa, G. Reagents,
methods, and libraries for bead-based sequencing. US Patent Application
11/345,979 (2005).
- Zon, G. Automated synthesis of phosphorus-sulfur analogs of nucleic
acids-25 years on. New J. Chem., 34, 795-804 (2010).
DOI: 10.1039/b9nj00577c
- Web references can alternatively be found at: http://tinyurl.com/livchemref
- Niedringhaus, T. P. et al. Landscape of next generation
sequencing methodologies. Anal. Chem. 83, 4327-4341
(2011). DOI: 10.1021/ac2010857
- Peterson T.W., Nam, S.J., Darby, A. J.P. Morgan Next Generation
Sequencing Survey, 12 May 2010. Available at: http://www.genomicslawreport.com/wp-content/uploads/2011/04/JP-Morgan-NGS-Report.pdf
10. Sales figures taken from Appendix 1 of this survey.
- Genome study solves twins' mystery condition, Nature News, 15 June
2011 DOI: 10.1038/news.2011.368. (This story has been featured on Good
Morning America, the Today show, the New York Times and about 240 other
publications worldwide.)
- Schulte, J.H. Deep sequencing reveals differential expression of
microRNAs of favorable versus unfavorable neuroblastoma. Nucl. Acids
Res., 38, 5919-5928 (2010). DOI:10.1093/NAR/gkq342
- Letter of corroboration available from the Director of Business
Development now at TriLink Biotech (details at: http://zon.trilinkbiotech.com/about-jerry-zon/)10.
Between 1999 and 2011 he was at Applied Biosystems (subsequently Life
Technologies) as detailed in reference 9 above.